Quesnel Lake Exploitation Analysis 2017
The suggested citation for this analytic report is:
Thorley, J.L. and Dalgarno, S. (2018) Quesnel Lake Exploitation Analysis 2017. A Poisson Consulting Analysis Report. URL: https://www.poissonconsulting.ca/f/942646731.
Background
Quesnel Lake supports a recreational fishery for large Bull Trout, Lake Trout and Rainbow Trout. To provide information on the natural and fishing mortality, trout were caught by angling and tagged with acoustic transmitters and/or reward tags.
Methods
Data Preparation
The outing, receiver deployment and fish capture and recapture information were provided by the Ministry of Forests, Lands and Natural Resource Operations and entered by Vicky Lipinski and databased by Gary Pavan.
The data were prepared for analysis using R version 3.4.4 (R Core Team 2018). Receivers were assumed to have a detection range of 500 m. Only individuals with a fork length (FL) \(\geq\) 450 mm, an acoustic tag life \(\geq\) 365 days (if acoustically tagged) and a $100 and $10 reward tags were included in the survival analysis.
Data Analysis
Hierarchical Bayesian models were fitted to the data using R version
3.4.4 (R Core Team 2018) and JAGS 4.2.0 (Plummer 2015) which
interfaced with each other via the jmbr package. For additional
information on hierarchical Bayesian modelling in the BUGS language, of
which JAGS uses a dialect, the reader is referred to Kery and Schaub (2011, 41–44).
Unless indicated otherwise, the Bayesian analyses used normal and uniform prior distributions that were vague in the sense that they did not constrain the posteriors (Kery and Schaub 2011, 36). The posterior distributions were estimated from 1500 Markov Chain Monte Carlo (MCMC) samples thinned from the second halves of 3 chains (Kery and Schaub 2011, 38–40). Model convergence was confirmed by ensuring that \(\hat{R} \leq 1.05\) (Kery and Schaub 2011, 40) and \(\textrm{ESS} \geq 150\) for each of the monitored parameters (Kery and Schaub 2011, 61). Where \(\hat{R}\) is the potential scale reduction factor and \(\textrm{ESS}\) is the effective sample size (Brooks et al. 2011).
The posterior distributions of the fixed (Kery and Schaub 2011, 75) parameters are summarised in terms of the point estimate, standard deviation (sd), the z-score, lower and upper 95% confidence/credible limits (CLs) and the p-value (Kery and Schaub 2011, 37, 42). The estimate is the median (50th percentile) of the MCMC samples, the z-score is \(\mathrm{mean}/\mathrm{sd}\) and the 95% CLs are the 2.5th and 97.5th percentiles. A p-value of 0.05 indicates that the lower or upper 95% CL is 0.
The results are displayed graphically by plotting the modeled relationships between particular variables and the response with the remaining variables held constant. In general, continuous and discrete fixed variables are held constant at their mean and first level values respectively while random variables are held constant at their typical values (expected values of the underlying hyperdistributions) (Kery and Schaub 2011, 77–82). Where informative the influence of particular variables is expressed in terms of the effect size (i.e., percent change in the response variable) with 95% CIs (Bradford, Korman, and Higgins 2005).
Model Descriptions
Survival
The natural mortality and exploitation were estimated using a Bayesian individual state-space survival model (Thorley and Andrusak 2017) with monthly intervals. The survival model incorporated natural and handling mortality, acoustic detection, inter-section movement, T-bar tag loss, recapture and reporting. In addition to assumptions 1 to 10, in Thorley and Andrusak (2017), the model also assumes that:
- The effect of handling on mortality lasts up to two months after capture.
- Annual T-bar tag-loss is between 1 and 30%.
- All recaptured fish are retained.
- Reporting of recaptured fish with one or more T-bar tags is between 90 and 100%.
Condition
The expected weight of fish of a given length were estimated from the data using a mass-length model (He et al. 2008). Key assumptions of the condition model include:
- The expected weight is allowed to vary with length and date.
- The residual variation in weight is log-normally distributed.
Yield-Per-Recruit
The optimal fishing mortality (to maximize number of individuals harvested) was calculated using a yield-per-recruit approach (Bison, O’Brien, and Martell 2003). Key assumptions include:
- The population is at equilibrium.
- All captured individuals are retained.
- There are no Allee effects.
- The life-history parameters are fixed.
Results
Templates
Survival
model{
bMortality ~ dnorm(-3, 3^-2)
bMortalityHandling ~ dnorm(0, 2^-2)
bTagLoss ~ dunif(1 - (1 - 0.01)^(1/12), 1 - (1 - 0.30)^(1/12))
bDetected ~ dunif(0, 1)
bMoved ~ dunif(0, 1)
bRecaptured ~ dunif(0, 1 - (1 - 0.50)^(1/12))
bReported ~ dunif(0.9, 1.00)
for (i in 1:nCapture){
logit(eMortality[i,PeriodCapture[i]]) <- bMortality + bMortalityHandling
eTagLoss[i,PeriodCapture[i]] <- bTagLoss
eDetected[i,PeriodCapture[i]] <- bDetected
eMoved[i,PeriodCapture[i]] <- bMoved
eRecaptured[i,PeriodCapture[i]] <- bRecaptured
eReported[i,PeriodCapture[i]] <- bReported
InLake[i,PeriodCapture[i]] <- 1
Alive[i,PeriodCapture[i]] ~ dbern(1-eMortality[i,PeriodCapture[i]])
TBarTag100[i,PeriodCapture[i]] ~ dbern(1-eTagLoss[i,PeriodCapture[i]])
TBarTag10[i,PeriodCapture[i]] ~ dbern(1-eTagLoss[i,PeriodCapture[i]])
Detected[i,PeriodCapture[i]] ~ dbern(Monitored[i,PeriodCapture[i]] * eDetected[i,PeriodCapture[i]])
Moved[i,PeriodCapture[i]] ~ dbern(Alive[i,PeriodCapture[i]] * Monitored[i,PeriodCapture[i]] * eMoved[i,PeriodCapture[i]])
Recaptured[i,PeriodCapture[i]] ~ dbern(Alive[i,PeriodCapture[i]] * eRecaptured[i,PeriodCapture[i]])
Reported[i,PeriodCapture[i]] ~ dbern(Recaptured[i,PeriodCapture[i]] * eReported[i,PeriodCapture[i]] * step(TBarTag100[i,PeriodCapture[i]] + TBarTag10[i,PeriodCapture[i]] - 1))
for(j in (PeriodCapture[i]+1):nPeriod) {
logit(eMortality[i,j]) <- bMortality + bMortalityHandling * step(PeriodCapture[i] - j + 2)
eTagLoss[i,j] <- bTagLoss
eDetected[i,j] <- bDetected
eMoved[i,j] <- bMoved
eRecaptured[i,j] <- bRecaptured
eReported[i,j] <- bReported
InLake[i,j] ~ dbern(InLake[i,j-1] * (1-Recaptured[i,j-1]))
Alive[i,j] ~ dbern(Alive[i,j-1] * (1-Recaptured[i,j-1]) * (1-eMortality[i,j-1]))
TBarTag100[i,j] ~ dbern(TBarTag100[i,j-1] * (1-Recaptured[i,j-1]) * (1-eTagLoss[i,j]))
TBarTag10[i,j] ~ dbern(TBarTag10[i,j-1] * (1-Recaptured[i,j-1]) * (1-eTagLoss[i,j]))
Detected[i,j] ~ dbern(InLake[i,j] * Monitored[i,j] * eDetected[i,j])
Moved[i,j] ~ dbern(Alive[i,j] * Monitored[i,j] * eMoved[i,j])
Recaptured[i,j] ~ dbern(Alive[i,j] * eRecaptured[i,j])
Reported[i,j] ~ dbern(Recaptured[i,j] * eReported[i,j] * step(TBarTag100[i,j] + TBarTag10[i,j] - 1))
}
}
..Template 1.
Condition
model {
bWeight ~ dnorm(0, 2^-2)
bWeightLength ~ dnorm(3, 2^-2)
bWeightDayte ~ dnorm(0, 2^-2)
sWeight ~ dnorm(0, 5^-2)
for(i in 1:length(Length)) {
eWeight[i] <- bWeight + bWeightDayte * Dayte[i] + bWeightLength * Length[i]
Weight[i] ~ dlnorm(eWeight[i], exp(sWeight)^-2)
}
W_p <- bWeightLength
W_500 <- exp(bWeight)
..Template 2.
Tables
Survival
Table 1. Parameter descriptions.
| Parameter | Description |
|---|---|
bDetected |
Logit monthly probability of detection if in-lake |
bMortality |
Logit monthly probability of dying of natural causes |
bMortalityHandling |
Effect of capture and handling on bMortality |
bMoved |
Logit monthly probability of being detected moving between sections if alive |
bRecaptured |
Logit monthly probability of being recaptured |
bReported |
Monthly probability of being reported if recaptured with one or more T-bar tags |
bTagLoss |
Monthly probability of loss for a single T-bar tag |
Bull Trout
Table 2. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bDetected | 0.4169320 | 0.0107617 | 38.750084 | 0.3959670 | 0.4382811 | 0.0007 |
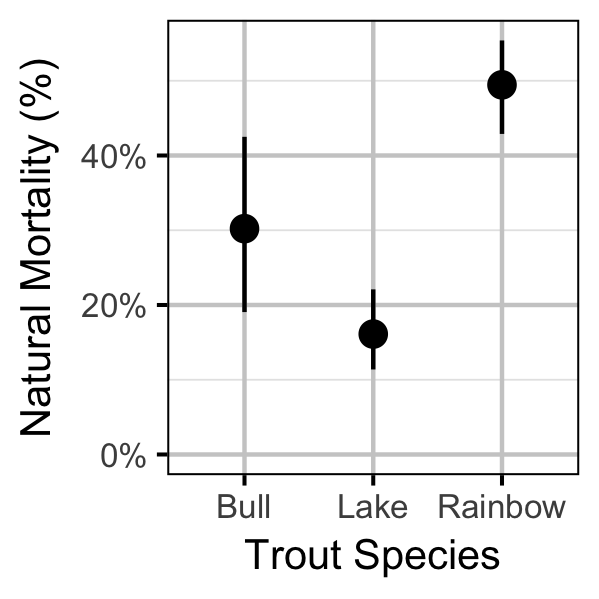
| bMortality | -3.4923604 | 0.2457833 | -14.271143 | -4.0301724 | -3.0534896 | 0.0007 |
| bMortalityHandling | 1.0727630 | 0.4133397 | 2.552433 | 0.2286481 | 1.8572029 | 0.0147 |
| bMoved | 0.1773531 | 0.0129811 | 13.699811 | 0.1540948 | 0.2041313 | 0.0007 |
| bRecaptured | 0.0048835 | 0.0019410 | 2.650326 | 0.0020804 | 0.0097071 | 0.0007 |
| bReported | 0.9572536 | 0.0289644 | 32.956158 | 0.9027288 | 0.9981692 | 0.0007 |
| bTagLoss | 0.0099161 | 0.0062226 | 1.754576 | 0.0021213 | 0.0259348 | 0.0007 |
Table 3. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 4675 | 7 | 3 | 500 | 500 | 53 | 1.085 | FALSE |
Lake Trout
Table 4. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bDetected | 0.7952307 | 0.0064009 | 124.219474 | 0.7818868 | 0.8076625 | 0.0007 |
| bMortality | -4.2166810 | 0.1889221 | -22.314892 | -4.5937970 | -3.8623131 | 0.0007 |
| bMortalityHandling | 0.9918517 | 0.3149927 | 3.130487 | 0.3507571 | 1.5607728 | 0.0013 |
| bMoved | 0.4540679 | 0.0094711 | 47.914867 | 0.4355635 | 0.4720405 | 0.0007 |
| bRecaptured | 0.0030575 | 0.0005579 | 5.508281 | 0.0020813 | 0.0042957 | 0.0007 |
| bReported | 0.9799874 | 0.0235677 | 41.294378 | 0.9128165 | 0.9992992 | 0.0007 |
| bTagLoss | 0.0013730 | 0.0008288 | 1.965480 | 0.0008621 | 0.0039681 | 0.0007 |
Table 5. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 24860 | 7 | 3 | 500 | 500 | 58 | 1.041 | FALSE |
Rainbow Trout
Table 6. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bDetected | 0.5603467 | 0.0068584 | 81.683821 | 0.5462199 | 0.5738301 | 0.0007 |
| bMortality | -2.8386594 | 0.0989192 | -28.732253 | -3.0410289 | -2.6652109 | 0.0007 |
| bMortalityHandling | -0.4992109 | 0.2245297 | -2.239697 | -0.9644613 | -0.0874545 | 0.0253 |
| bMoved | 0.7066050 | 0.0085314 | 82.812592 | 0.6899037 | 0.7224901 | 0.0007 |
| bRecaptured | 0.0125388 | 0.0017724 | 7.164344 | 0.0096511 | 0.0163317 | 0.0007 |
| bReported | 0.9871643 | 0.0165066 | 59.497222 | 0.9382342 | 0.9996264 | 0.0007 |
| bTagLoss | 0.0033280 | 0.0016213 | 2.206780 | 0.0011467 | 0.0076593 | 0.0007 |
Table 7. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 20185 | 7 | 3 | 500 | 500 | 99 | 1.06 | FALSE |
Condition
Table 8. Parameter descriptions.
| Parameter | Description |
|---|---|
bWeight |
Intercept of eWeight |
bWeightDayte |
Effect of Dayte on bWeight |
bWeightLength |
Intercept of effect of Length on bWeight |
Dayte |
Standardised day of year of capture |
eWeight |
Log expected Weight |
Length |
Log-transformed and centered on 500mm fork length |
sWeight |
Log standard deviation of residual variation in log Weight |
W500 |
Weight at 500 mm (kg) |
Weight |
Mass (kg) |
Wp |
Weight power term |
Bull Trout
Table 9. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bWeight | 0.2470391 | 0.0463567 | 5.369646 | 0.1586015 | 0.3390328 | 0.0007 |
| bWeightDayte | 0.0294158 | 0.0275804 | 1.062235 | -0.0265500 | 0.0806554 | 0.2733 |
| bWeightLength | 3.2939603 | 0.2086140 | 15.778112 | 2.8908608 | 3.7156016 | 0.0007 |
| sWeight | -1.6326989 | 0.0995449 | -16.353686 | -1.8212933 | -1.4198944 | 0.0007 |
Table 10. Derived coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| W_500 | 1.280229 | 0.0596763 | 21.51637 | 1.171871 | 1.403590 | 7e-04 |
| W_p | 3.293960 | 0.2086140 | 15.77811 | 2.890861 | 3.715602 | 7e-04 |
Table 11. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 56 | 4 | 3 | 500 | 1 | 186 | 1.007 | TRUE |
Lake Trout
Table 12. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bWeight | 0.2956832 | 0.0173038 | 17.112181 | 0.2613740 | 0.3298025 | 0.0007 |
| bWeightDayte | 0.0284693 | 0.0085946 | 3.319793 | 0.0111546 | 0.0460031 | 0.0013 |
| bWeightLength | 3.1414125 | 0.0773853 | 40.564267 | 2.9889160 | 3.2848537 | 0.0007 |
| sWeight | -1.9593239 | 0.0438079 | -44.716025 | -2.0389539 | -1.8703952 | 0.0007 |
Table 13. Derived coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| W_500 | 1.344044 | 0.0232833 | 57.75877 | 1.298713 | 1.390693 | 7e-04 |
| W_p | 3.141412 | 0.0773853 | 40.56427 | 2.988916 | 3.284854 | 7e-04 |
Table 14. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 262 | 4 | 3 | 500 | 1 | 189 | 1.012 | TRUE |
Rainbow Trout
Table 15. Model coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| bWeight | 0.4093824 | 0.0162670 | 25.175583 | 0.3768944 | 0.4425569 | 7e-04 |
| bWeightDayte | 0.0578334 | 0.0108988 | 5.294111 | 0.0355493 | 0.0797535 | 7e-04 |
| bWeightLength | 3.0596462 | 0.0772934 | 39.581225 | 2.9060432 | 3.2092630 | 7e-04 |
| sWeight | -1.7163188 | 0.0433542 | -39.538284 | -1.7961453 | -1.6295262 | 7e-04 |
Table 16. Derived coefficients.
| term | estimate | sd | zscore | lower | upper | pvalue |
|---|---|---|---|---|---|---|
| W_500 | 1.505888 | 0.0245204 | 61.43093 | 1.457750 | 1.556682 | 7e-04 |
| W_p | 3.059646 | 0.0772934 | 39.58122 | 2.906043 | 3.209263 | 7e-04 |
Table 17. Model summary.
| n | K | nchains | niters | nthin | ess | rhat | converged |
|---|---|---|---|---|---|---|---|
| 277 | 4 | 3 | 500 | 1 | 246 | 1.017 | TRUE |
Yield-Per-Recruit
Table 18. Assumed life-history parameters values in yield-per-recruit calculation by trout species.
| Parameter | Symbol | Bull | Lake | Rainbow |
|---|---|---|---|---|
| Asymptotic length (mm) | \(L_{\infty}\) | 1000.00 | 1000.00 | 1000.0 |
| Growth constant (y-1) | \(k\) | 0.13 | 0.15 | 0.2 |
| Weight at 500 mm (kg) | \(W_{500}\) | 1.30 | 1.40 | 1.6 |
| Weight power | \(W_{p}\) | 3.40 | 3.10 | 3.0 |
| Length at maturity (mm) | \(L_{m}\) | 500.00 | 500.00 | 500.0 |
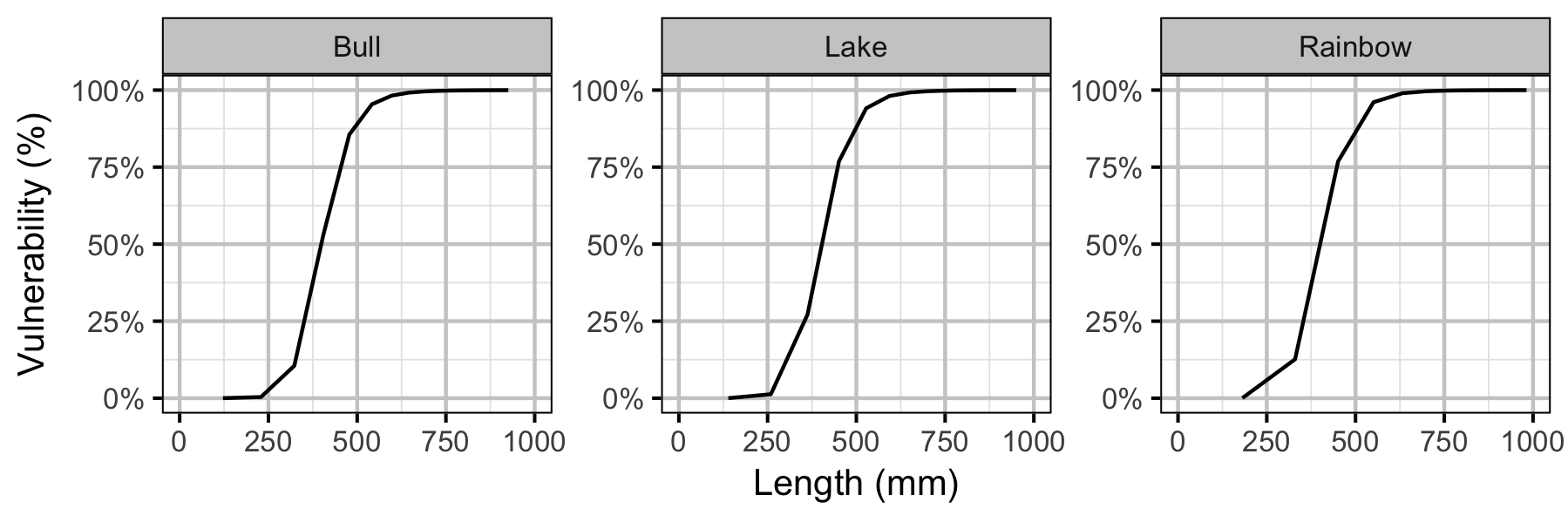
| Length at which 50% vulnerable to angling (mm) | \(L_{v}\) | 400.00 | 400.00 | 400.0 |
| Vulnerability power | \(V_{p}\) | 10.00 | 10.00 | 10.0 |
| Maximum annual reproductive rate | \(R_{k}\) | 3.80 | 2.50 | 6.0 |
Table 19. Yield-per-recruit calculations.
| Parameter | Symbol | Relationship |
|---|---|---|
| Annual interval fishing mortality rate | \(U\) | \(\text{Optimized by yield-per-recruit analysis}\) |
| Annual interval natural mortality rate | \(V\) | \(\text{From survival analysis}\) |
| Annual instantaneous fishing mortality rate | \(F\) | \(-\text{log}(1-U)\) |
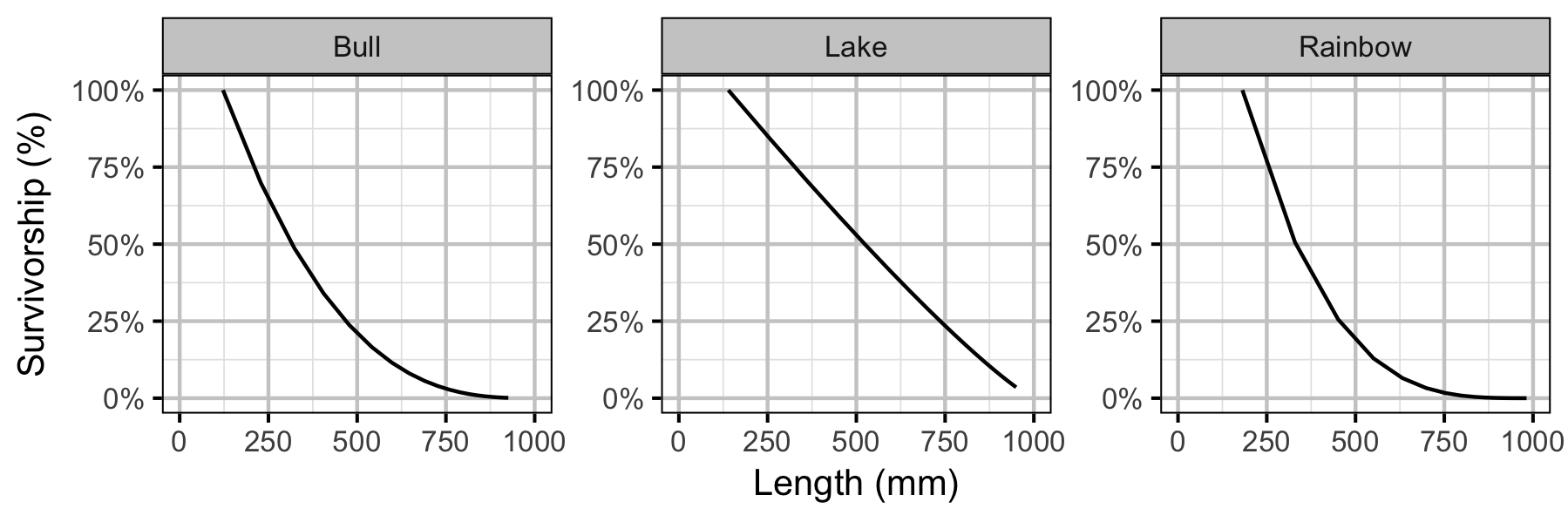
| Annual instantaneous natural mortality rate | \(M\) | \(-\text{log}(1-V)\) |
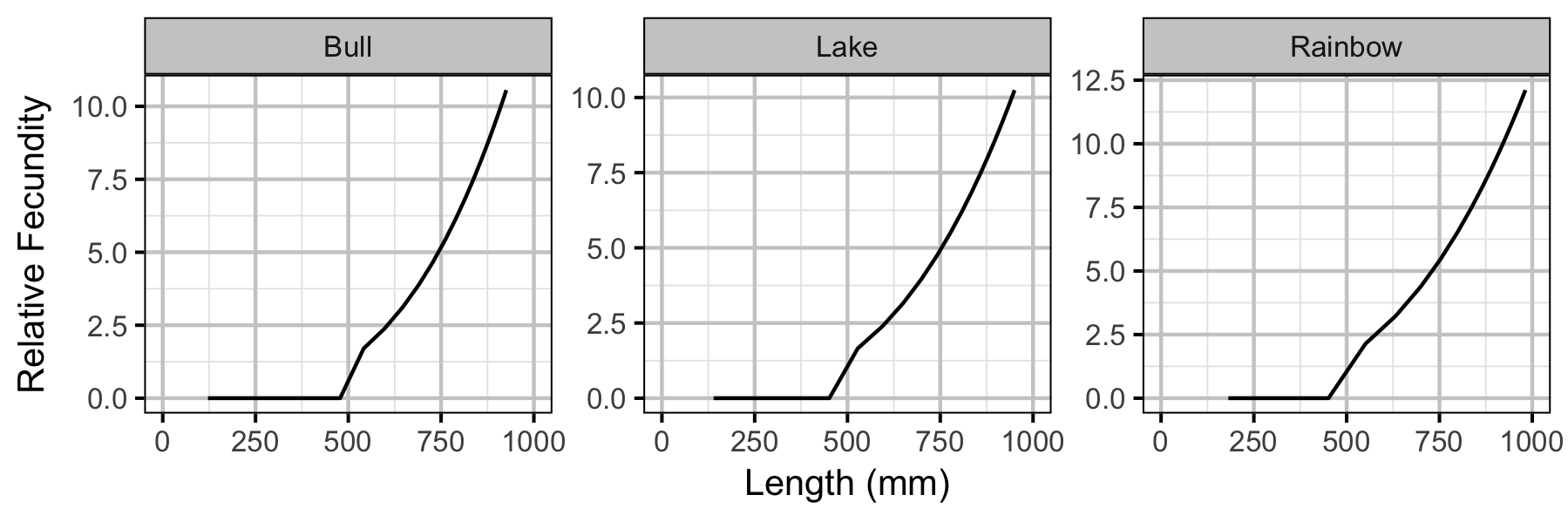
| Length at age a (mm) | \(L_{a}\) | \(L_{\infty}(1-\exp(-k \cdot a))\) |
| Weight at age a (kg) | \(W_{a}\) | \((W_{500} / 500^{W_p}) \cdot L_{a}^{W_p}\) |
| Relative fecundity at age a | \(F_{a}\) | \(W_{a} \: \text{if} \: (L_{a} \geq L_{m}) \: \mathrm{otherwise}\: 0\) |
| Vulnerability to fishing at age a | \(V_{a}\) | \(L_{a}^{V_p}/(L_{v}^{V_p} + L_{a}^{V_p})\) |
| Unfished survivorship to age a | \(\lambda_{a}\) | \(\exp(-M)^{a-1}\) |
| Fished survivorship to age a | \(\lambda'_{a}\) | \(\exp(-M+F \cdot V_a)^{a-1}\) |
| Unfished relative fecundity per age-1 recruit | \(\phi_{0}\) | \(\sum(\lambda_{a} \cdot F_{a})\) |
| Fished relative fecundity per age-1 recruit | \(\phi'_{0}\) | \(\sum(\lambda'_{a} \cdot F_{a})\) |
| Unfished age-1 recruits as percent of total unfished fish population (TUFP) (%) | \(R_{0}\) | \(100/\sum\lambda_{a}\) |
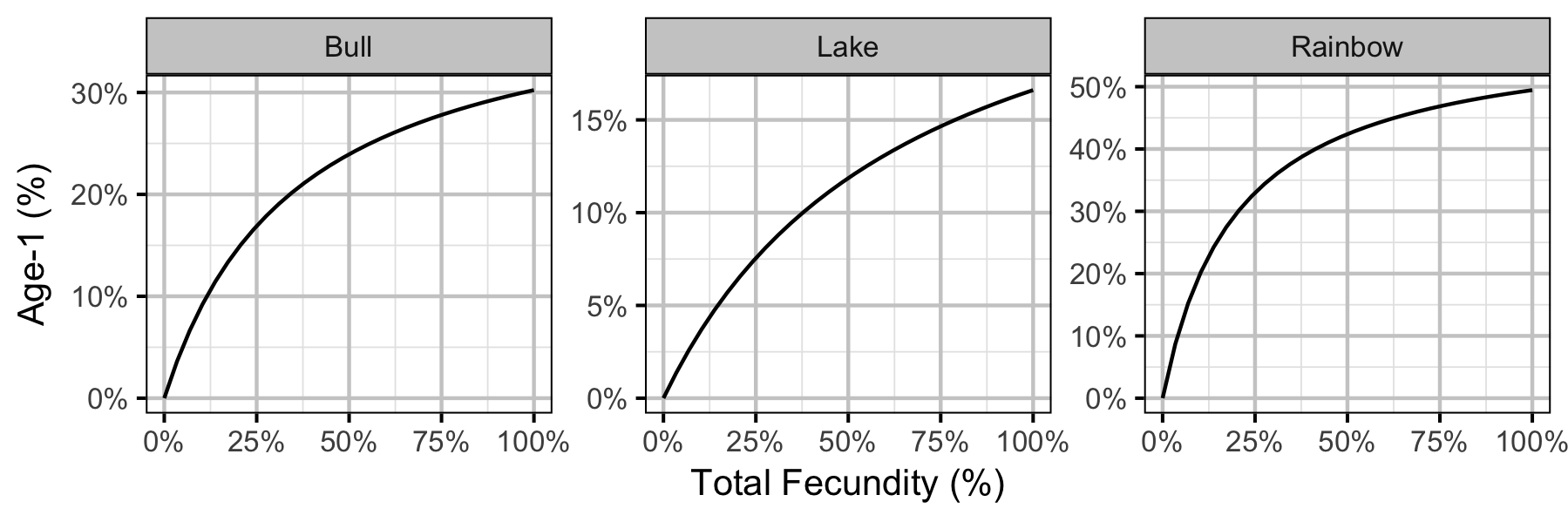
| Stock productivity at low density | \(\alpha\) | \(R_{k} \cdot \phi^{-1}\) |
| Stock density dependence | \(\beta\) | \((R_{k} - 1)/(R_{0} \cdot \phi)\) |
| Fished age-1 recruits as percent of TUFP (%) | \(R'_{0}\) | \((\alpha \cdot \phi'_{0} - 1)/(\beta \cdot \phi'_{0})\) |
| Equilibrium yield as percent of TUFP (%) | \(Y_{e}\) | \(U \cdot R'_{0} \cdot \sum(\lambda'_{a} \cdot V_{a})\) |
Recapture
Table 20.
| Species | Year | Captured | Reported |
|---|---|---|---|
| Bull Trout | 2013 | 23 | 0 |
| Bull Trout | 2014 | 16 | 0 |
| Bull Trout | 2015 | 13 | 2 |
| Bull Trout | 2016 | 24 | 1 |
| Bull Trout | 2017 | 9 | 3 |
| Lake Trout | 2013 | 101 | 1 |
| Lake Trout | 2014 | 182 | 9 |
| Lake Trout | 2015 | 74 | 2 |
| Lake Trout | 2016 | 59 | 9 |
| Lake Trout | 2017 | 36 | 11 |
| Rainbow Trout | 2013 | 55 | 1 |
| Rainbow Trout | 2014 | 53 | 13 |
| Rainbow Trout | 2015 | 55 | 6 |
| Rainbow Trout | 2016 | 140 | 18 |
| Rainbow Trout | 2017 | 64 | 13 |
Figures
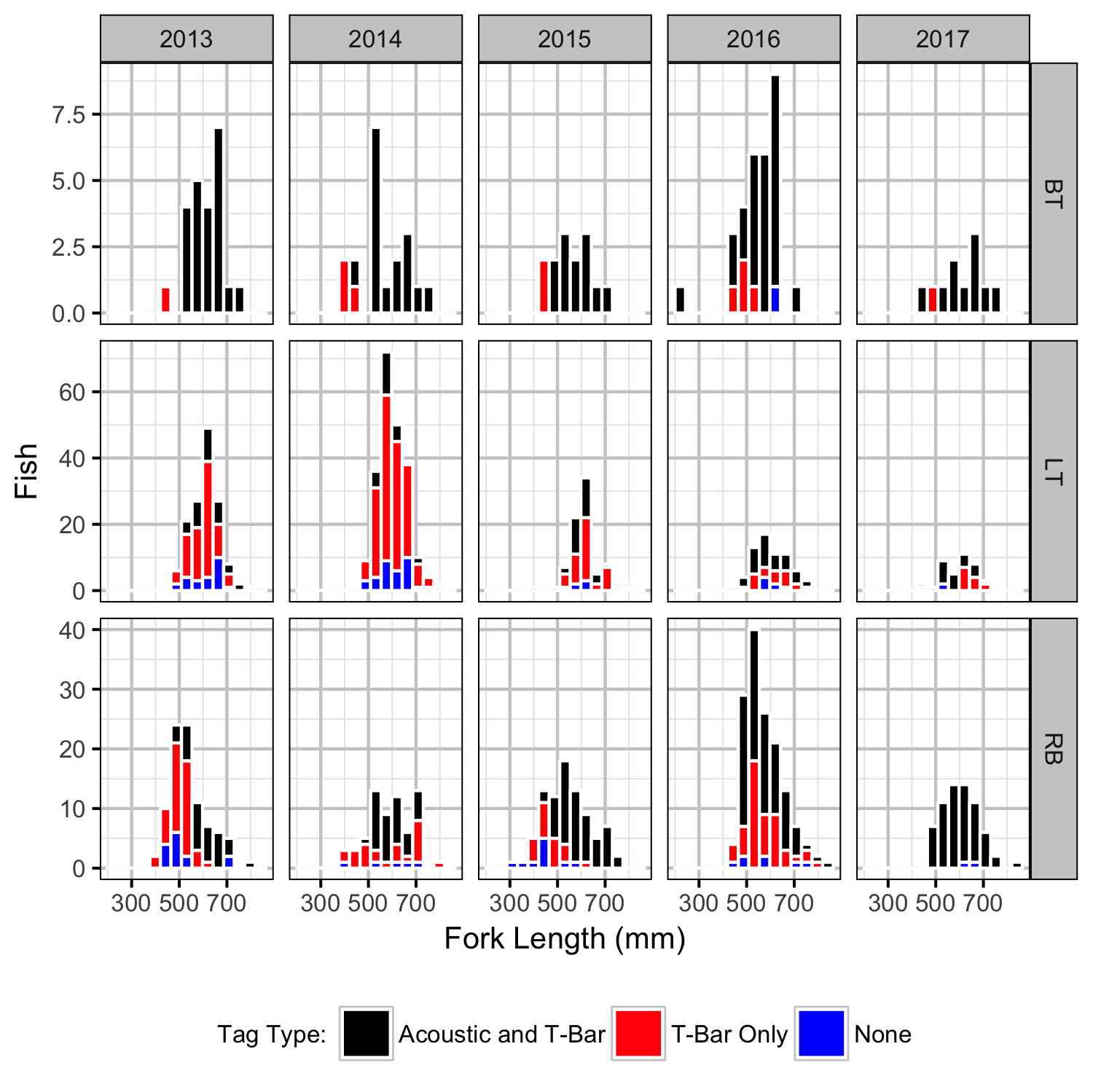
Capture
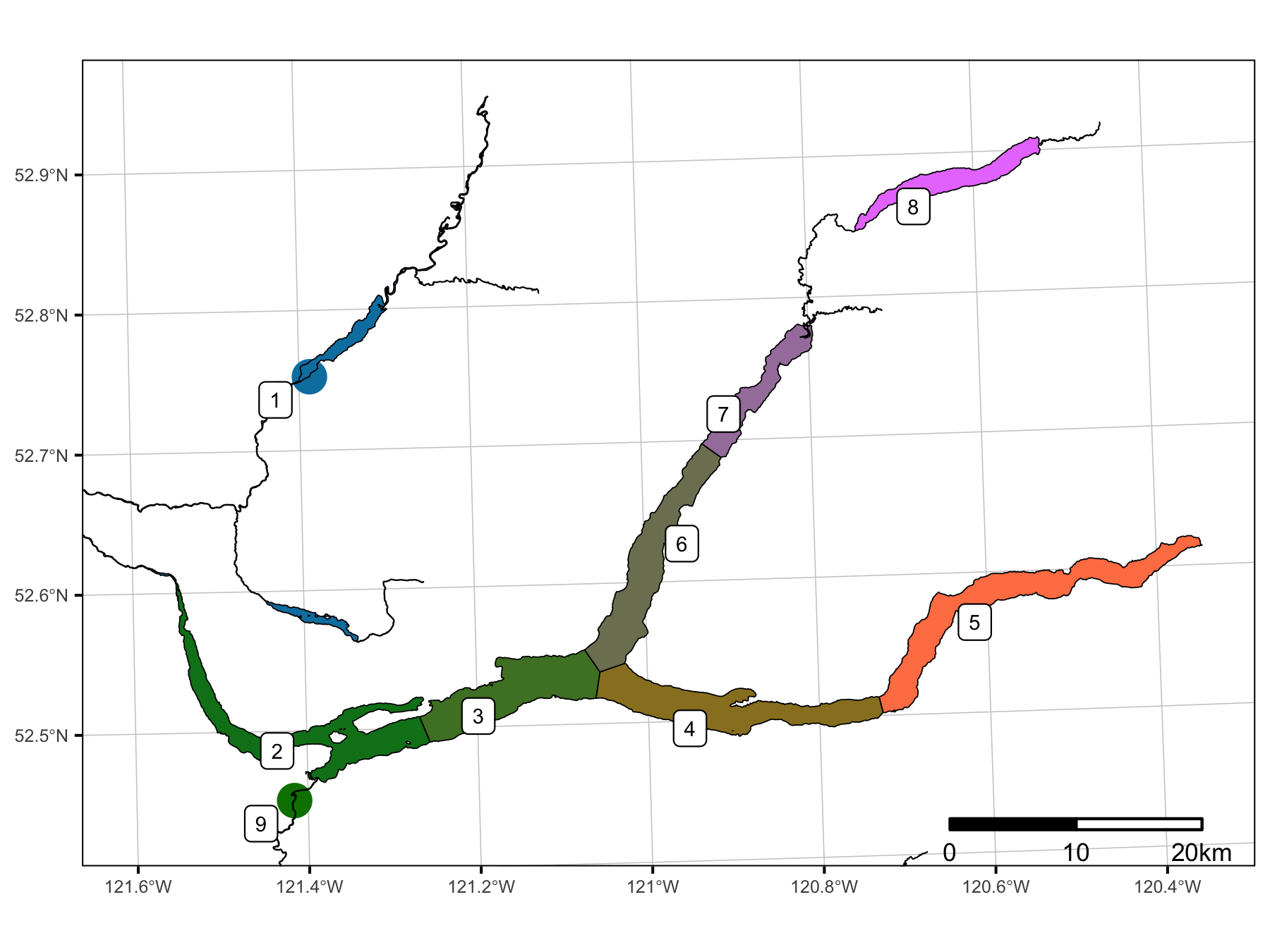
Sections
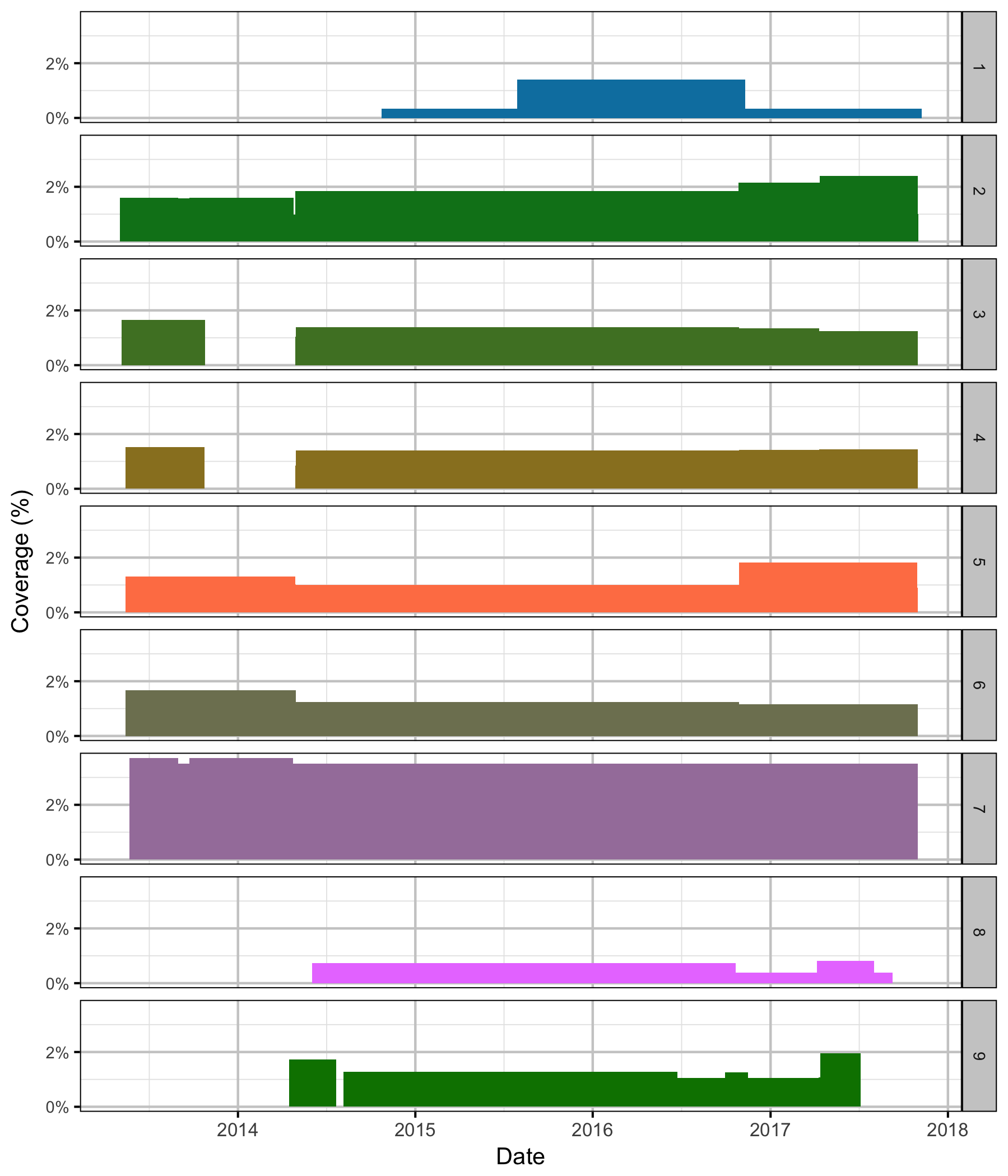
Coverage
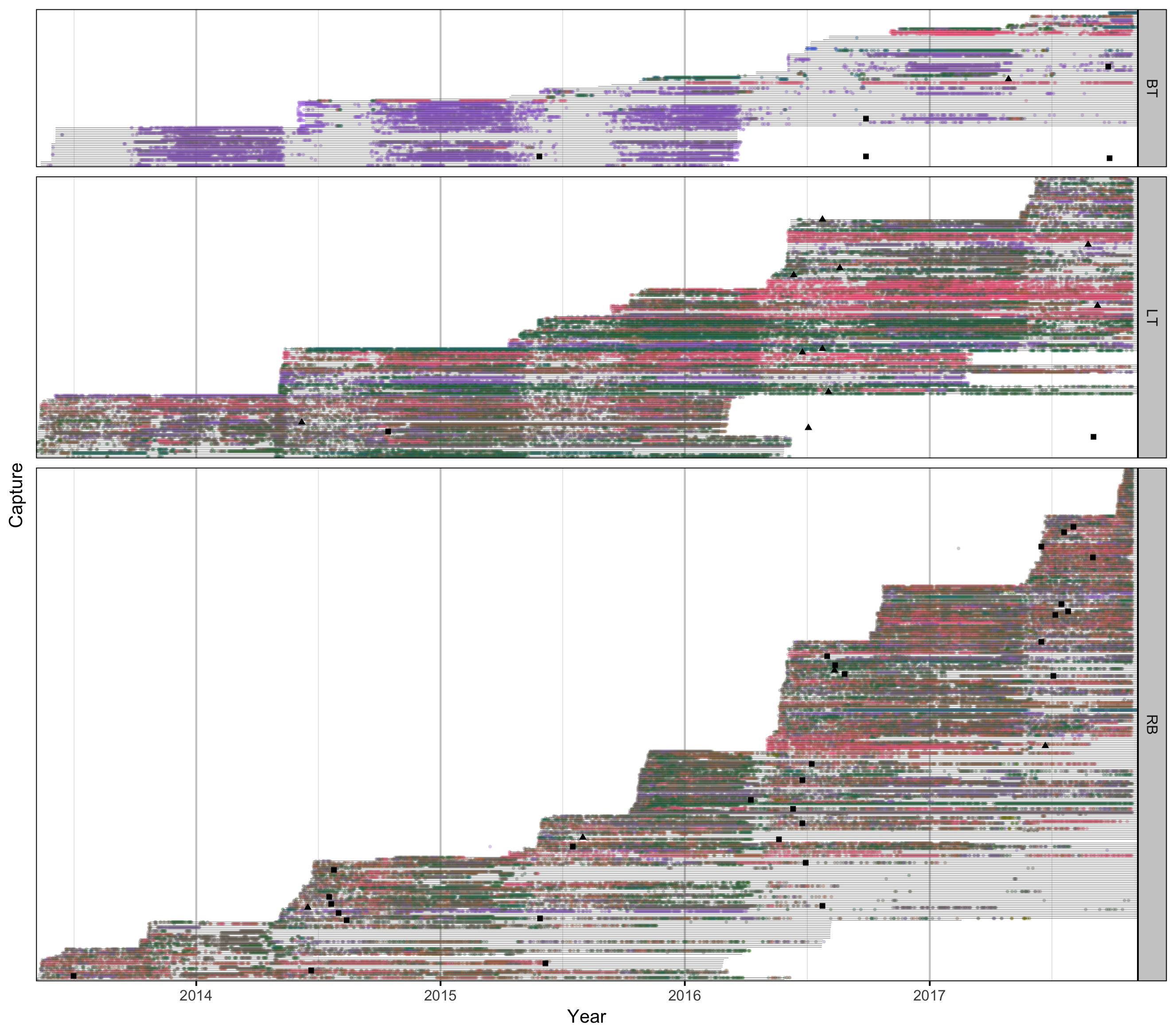
Detections
Section Use
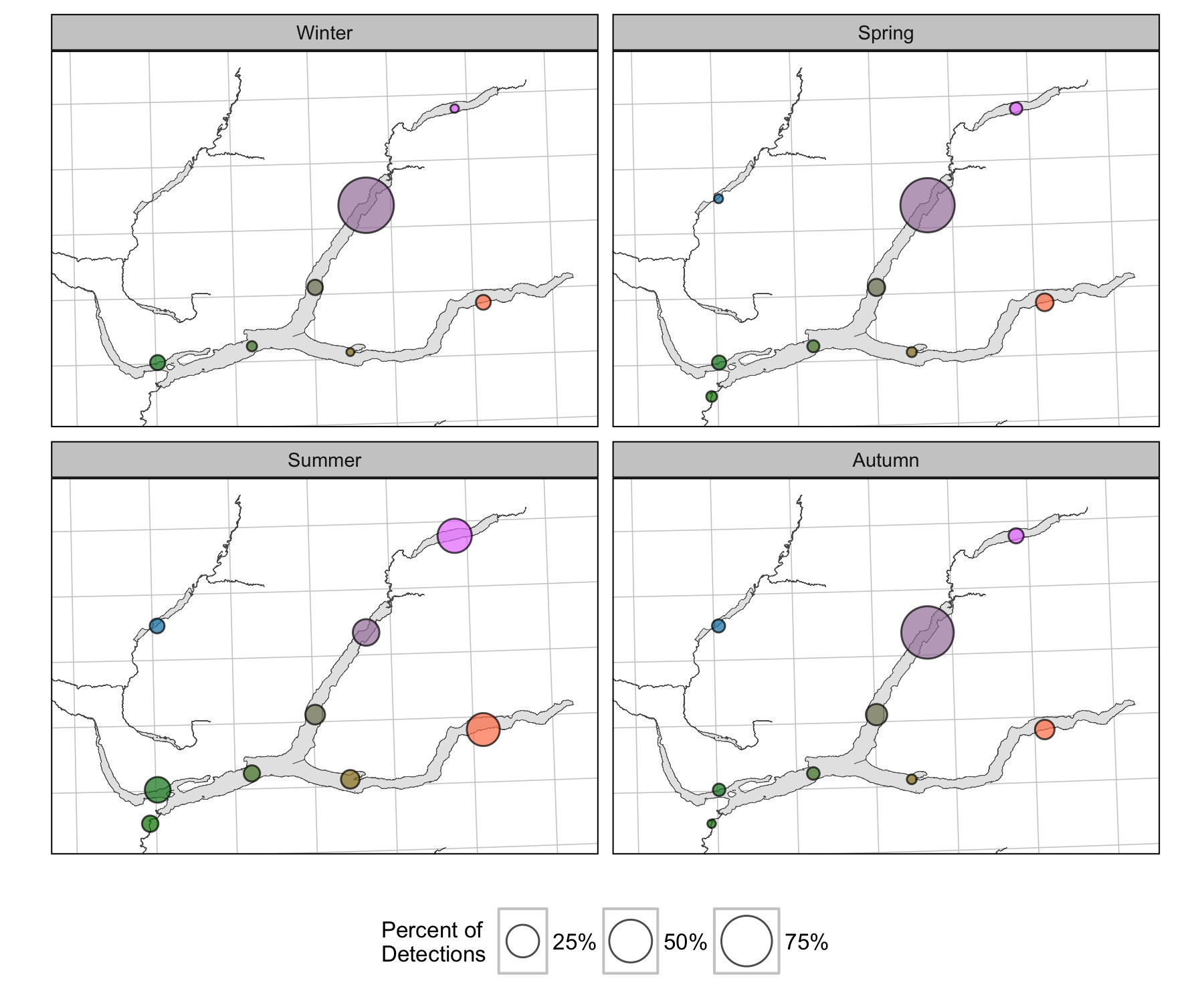
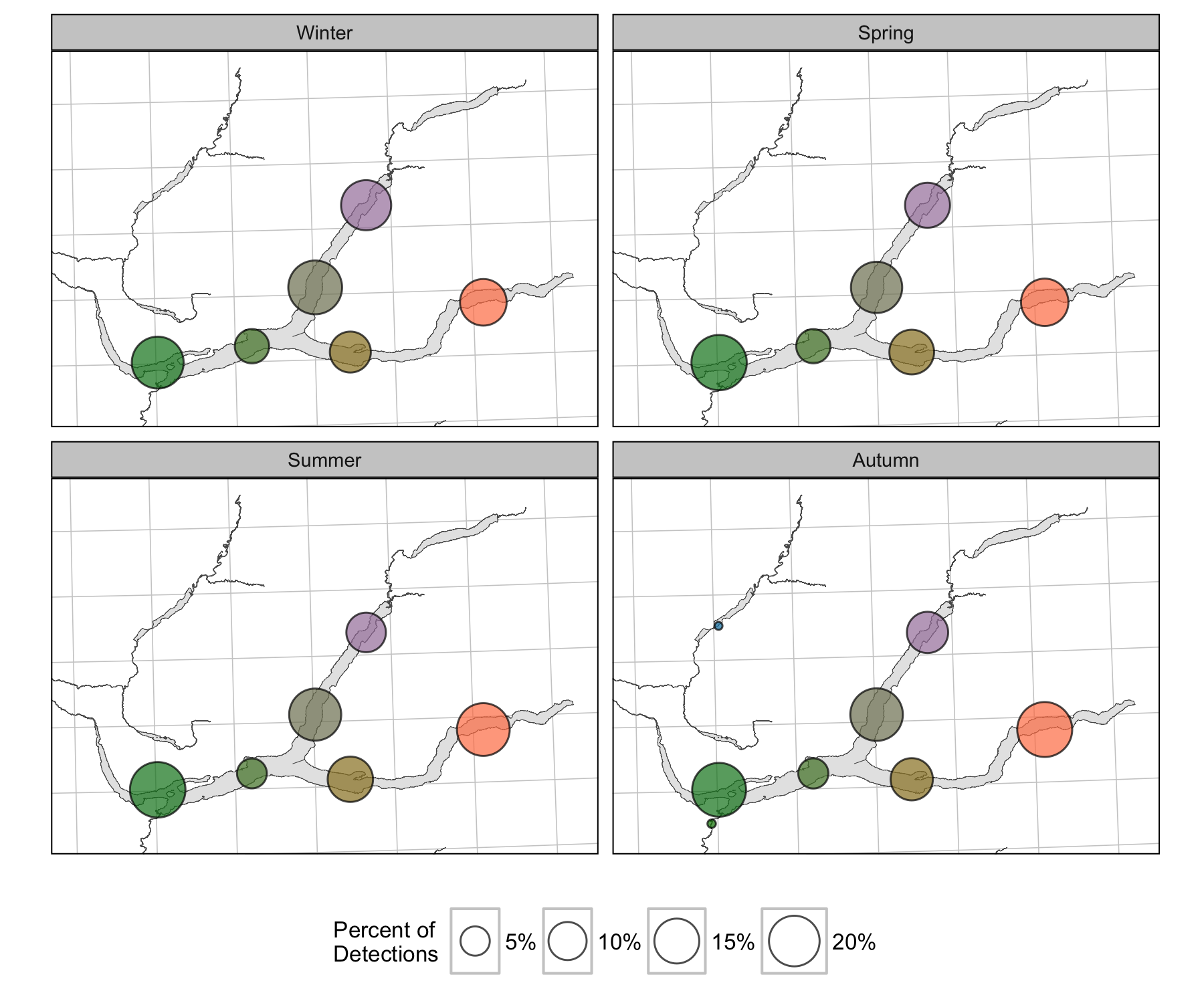
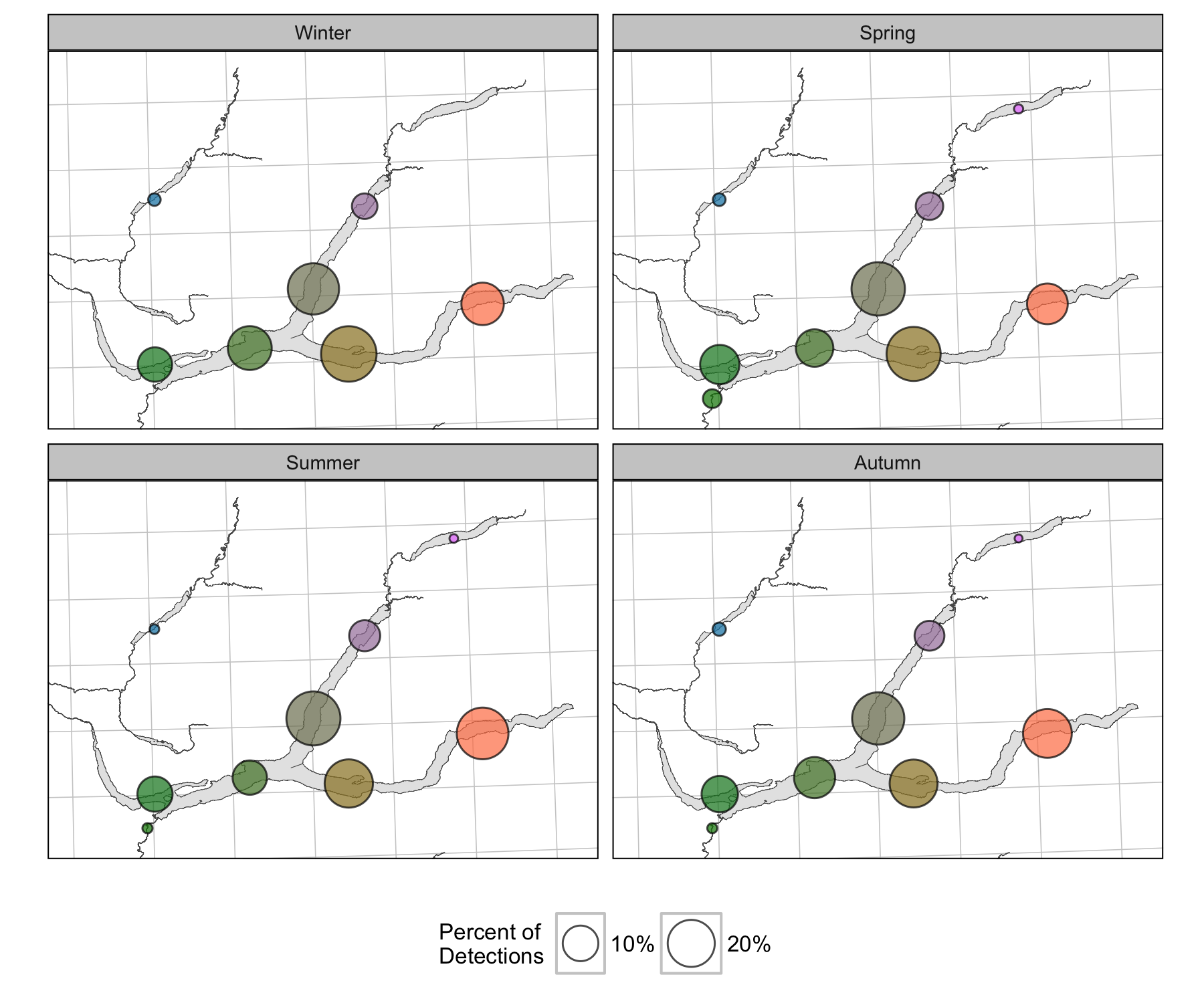
Survival
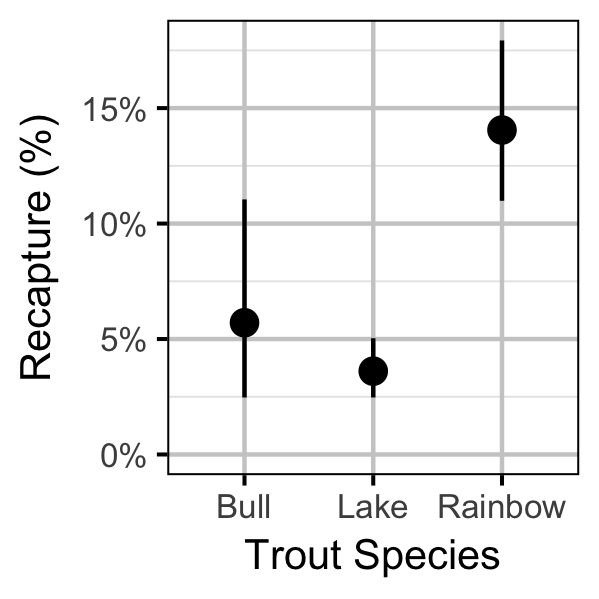
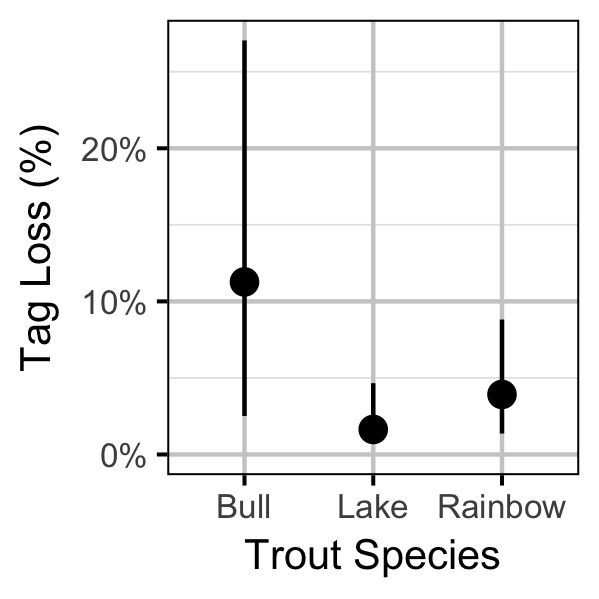
Yield-Per-Recruit
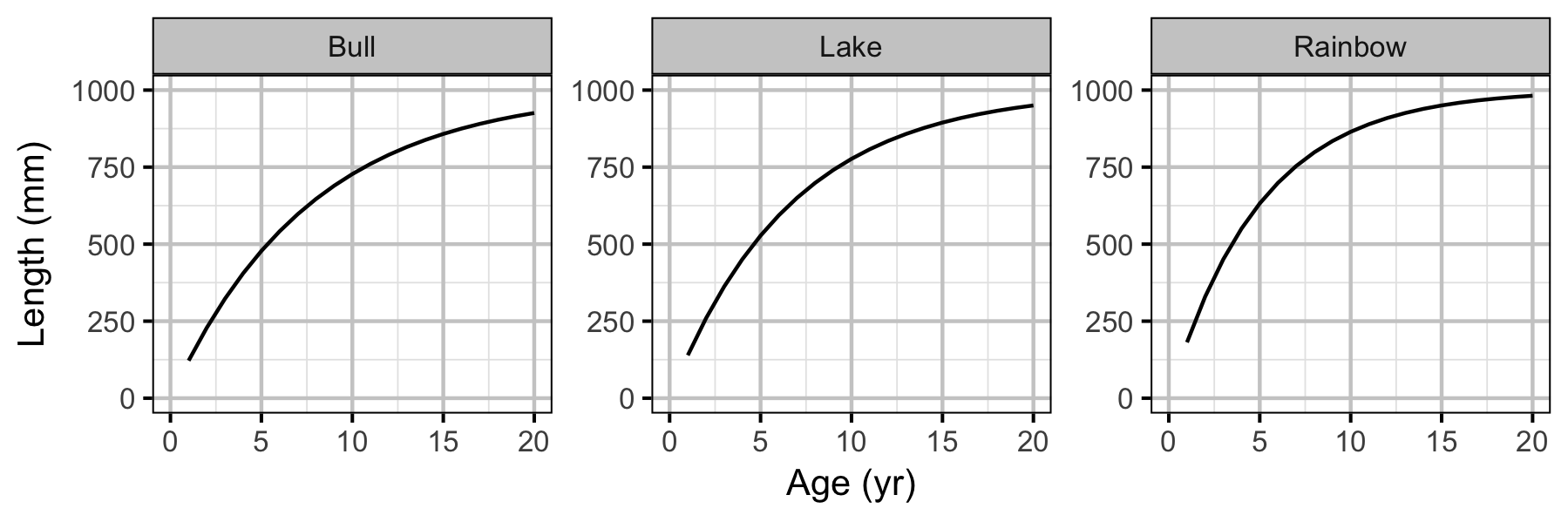
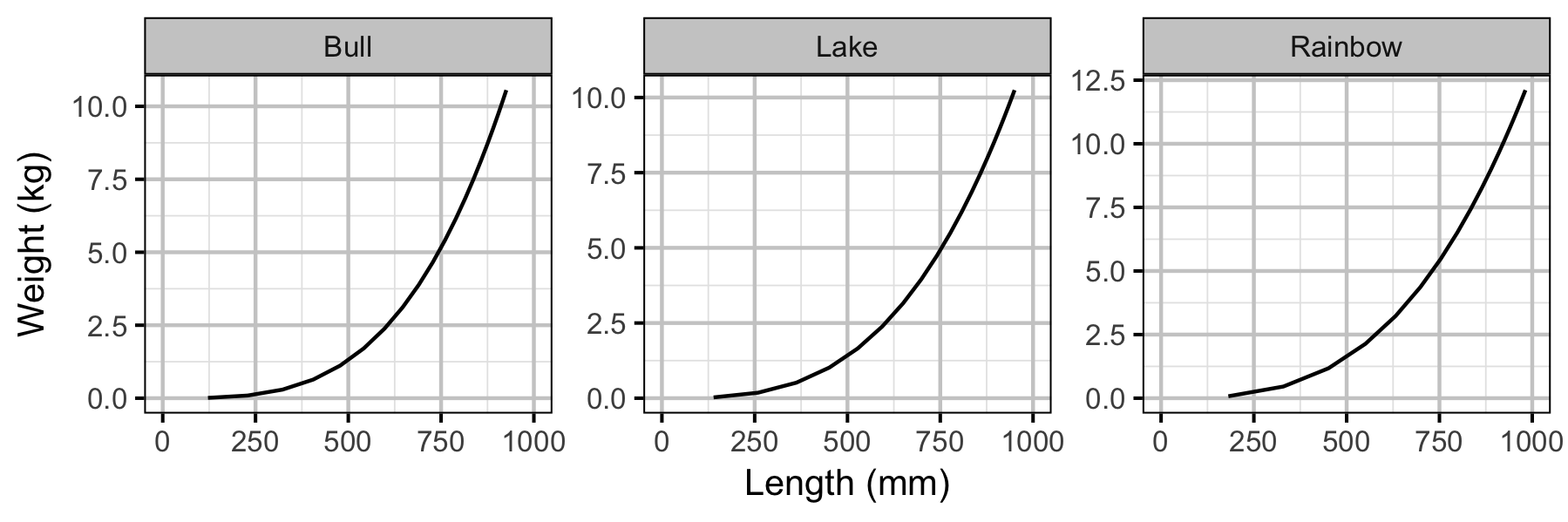
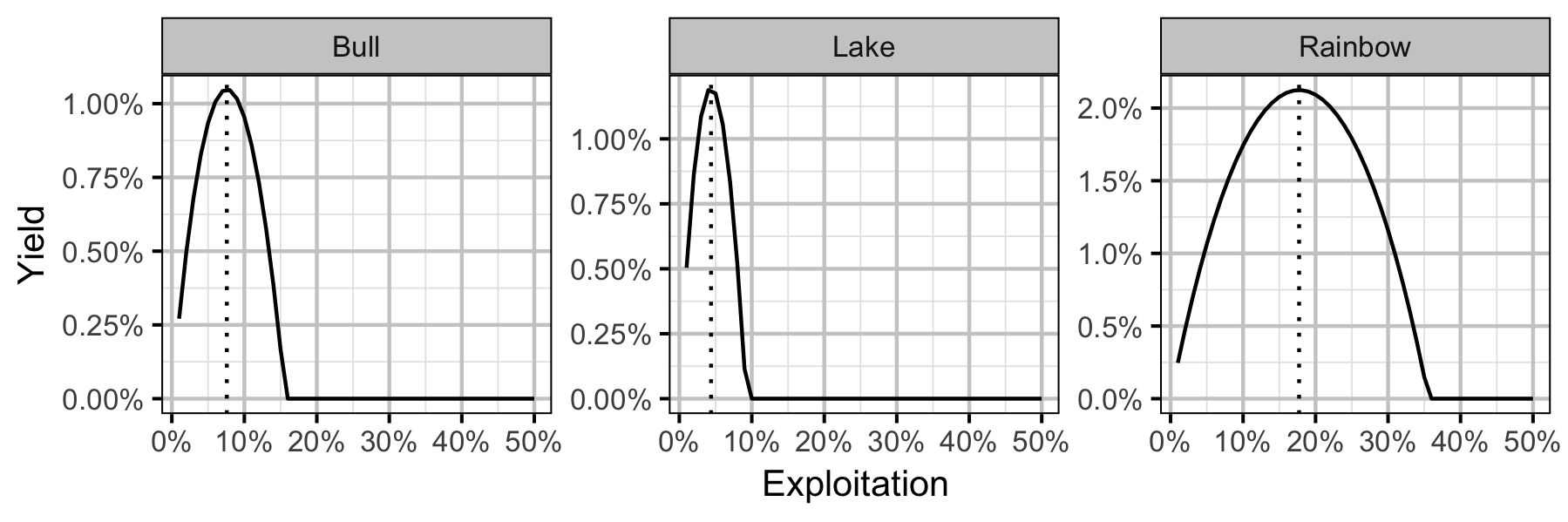
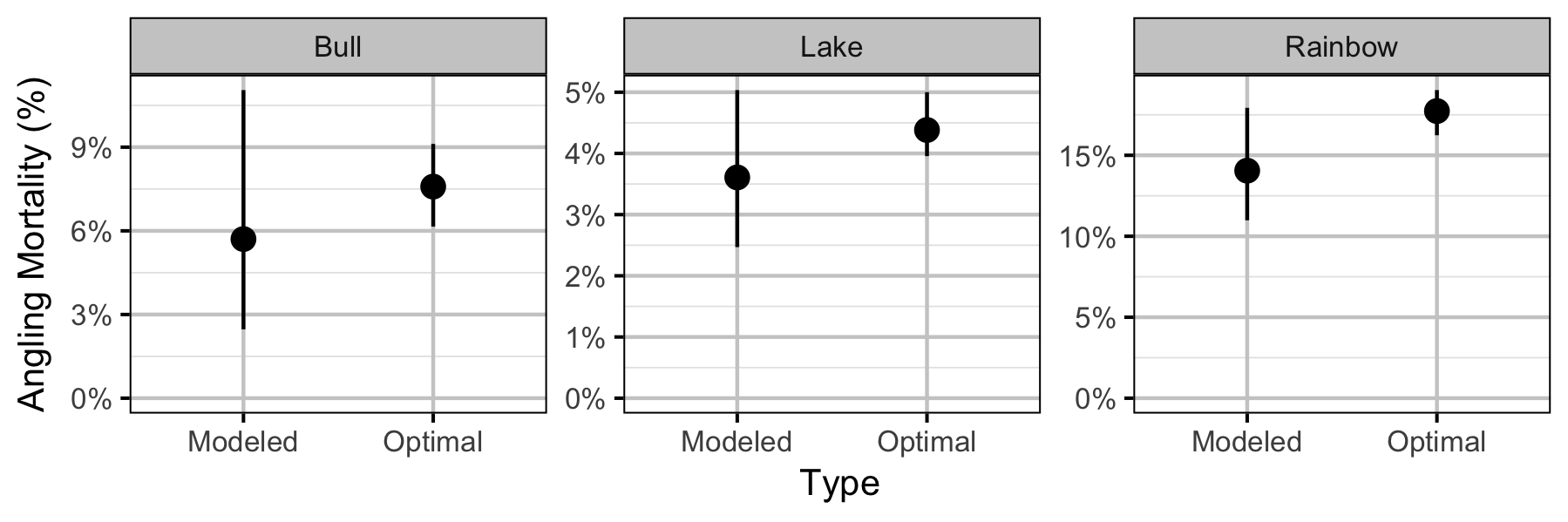
Conclusions
Recommendations
Recommendations include:
- Develop webpage animating individual fish movements.
- Incorporate recaptures by research crew into survival model.
- Add growth component to survival model to estimate growth parameters and adjust lengths.
- Explore seasonal, annual and length-based variation in natural and fishing mortality.
- Review life-history parameters, in particular \(R_k\), used in the yield-per-recruit calculations.
Acknowledgements
The organisations and individuals whose contributions have made this analysis report possible include:
- Quesnel Lake anglers for reporting their catches
- Habitat Conservation Trust Foundation (HCTF) and the anglers, hunters, trappers and guides who contribute to the Trust
- Ministry of Forests, Lands and Natural Resource Operations (MFLNRO)
- Lee Williston
- Greg Andrusak
- Mike Ramsay
- Reel Adventures
- Kerry Reed
- Vicky Lipinski
References
Bison, Robert, David O’Brien, and Steven J. D. Martell. 2003. “An Analysis of Sustainable Fishing Options for Adams Lake Bull Trout Using Life History and Telemetry Data.” Kamloops, B.C.: BC Ministry of Water Land; Air Protection.
Bradford, Michael J, Josh Korman, and Paul S Higgins. 2005. “Using Confidence Intervals to Estimate the Response of Salmon Populations (Oncorhynchus Spp.) to Experimental Habitat Alterations.” Canadian Journal of Fisheries and Aquatic Sciences 62 (12): 2716–26. https://doi.org/10.1139/f05-179.
Brooks, Steve, Andrew Gelman, Galin L. Jones, and Xiao-Li Meng, eds. 2011. Handbook for Markov Chain Monte Carlo. Boca Raton: Taylor & Francis.
He, Ji X., James R. Bence, James E. Johnson, David F. Clapp, and Mark P. Ebener. 2008. “Modeling Variation in Mass-Length Relations and Condition Indices of Lake Trout and Chinook Salmon in Lake Huron: A Hierarchical Bayesian Approach.” Transactions of the American Fisheries Society 137 (3): 801–17. https://doi.org/10.1577/T07-012.1.
Kery, Marc, and Michael Schaub. 2011. Bayesian Population Analysis Using WinBUGS : A Hierarchical Perspective. Boston: Academic Press. http://www.vogelwarte.ch/bpa.html.
Plummer, Martyn. 2015. “JAGS Version 4.0.1 User Manual.” http://sourceforge.net/projects/mcmc-jags/files/Manuals/4.x/.
R Core Team. 2018. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. https://www.R-project.org/.
Thorley, Joseph L., and Greg F. Andrusak. 2017. “The Fishing and Natural Mortality of Large, Piscivorous Bull Trout and Rainbow Trout in Kootenay Lake, British Columbia (2008–2013).” PeerJ 5 (January): e2874. https://doi.org/10.7717/peerj.2874.